This article will discuss the basic theory and relevant code examples for different clustering algorithms.
Code for different algorithms and data used are given in github.
Note – Make sure to point the code to the right directory in your setup
Clustering – Unsupervised learning algorithm
- It uses unlabeled training data X to separate it into y clusters (classes) using some similarity measure (distance, density, etc)
- Data points similar (close) to each other are assigned to one cluster
- It is unlike supervised algorithms that require labeled data to learn from in order to build models to make predictions
- It is a multi-objective optimization algorithm that uses an iterative process to find the best fit clusters for the training data
- A new unlabeled observation can then be assigned to the right cluster based on the distance metric used
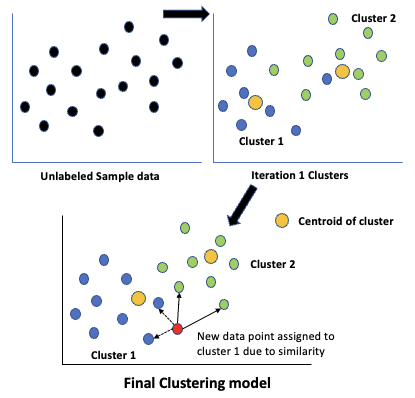
- In the figure shown, the clustering algorithm starts with the unlabeled sample data and performs multiple iterations of
- Splitting the data into target number of clusters and calculating their centroid
- In the next iteration it reassigns the data to clusters using the previous iterations centroids and calculates the new centroid, repeating this till an optimal value is reached
- The splitting, no of iterations, centroid calculation, etc depend on the algorithm used and its defined parameters
- No of clusters are generally user defined or estimated by the algorithm
Metrics:
- Silhouette Coefficient – defines how well defined and dense the clusters are. Useful when actual y values are unknown. Between (-1,1). Closer to 1 better
- Adjusted Rand index – proportional to the number of sample pairs with same labels for predicted and actual y labels. Between (0,1). Closer to 1 better
- Homogeneity – each cluster contains only members of a single class. Between (0,1). Closer to 1 better
- Completeness – all members of a given class are assigned to the same cluster. Between (0,1). Closer to 1 better
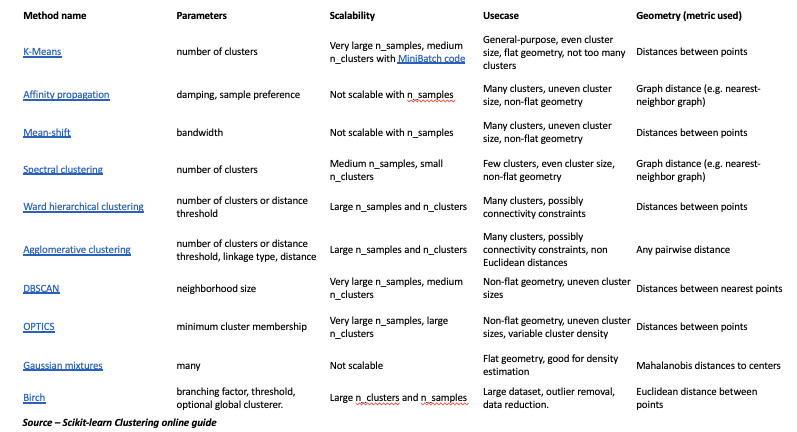
- Different clustering algorithms depend on the type of distance metric used. Some common one’s are discussed Useful website – https://scikit-learn.org/stable/modules/clustering.html
K-Means
- Centroid based K-Means clustering algorithm separates the n samples of X into k clusters by minimizing the squared distance of the data point from the cluster mean C (centroid)
- Centroids are selected so that the within-cluster-sum-of-squares WCSS (inertia) is minimized
- The equation to be solved is
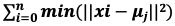 . μj ∈ C, is the mean of each cluster (j = 1 to k)
. μj ∈ C, is the mean of each cluster (j = 1 to k) - The algorithm works by first randomly initializing k, then repeats the below steps till the threshold is reached
- Assigns each sample xi to the nearest centroid μj
- It then creates new centroids by taking the mean of samples assigned to each previous centroid
- Difference between the mean of the old and new centroids is compared to the threshold
- These 3 steps are repeated till the threshold is reached
- As k is randomly fixed at the start, it is a drawback as the algorithm has to be repeated for different values
- To over come this random initialization issue the Elbow method and K-Means++ initialization are used
- The Elbow method gives a better initial value for k, by plotting the explained variance as a function of the no of clusters
- The ‘elbow’ or ‘knee’ of the curve acts as the cut-off point for k for diminishing returns
- The point where adding more clusters does not improve the model, cost vs benefit)
- K-means++ initialization further improves the results by initializing the centroids to be distant from each other
- K means algorithms assume that the clusters are convex shaped (outward curve, all parts pointing inwards. Eg: rugby ball)
- The code for plotting the elbow curve to determine initial k is given in the example
- It is determined by using the inertia_ attribute of the model
- It is the Sum of squared distances of samples to their closest cluster centre (WCSS)
- Results NBA rookie stats to predict if a player will last 5 years in league as 2 clusters are given below
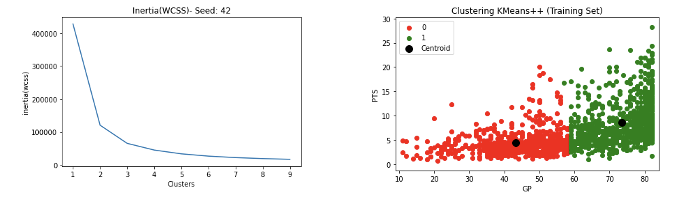
- Results for Kaggle Tortuga Code dataset to predict different students’ developer category as 6 clusters are given below
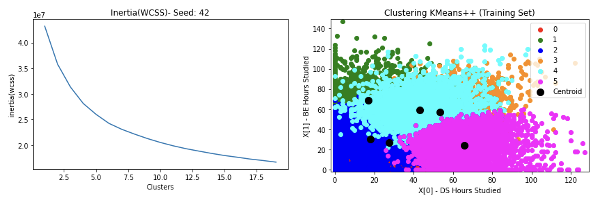
Hierarchical
- Connectivity based Hierarchical clustering algorithms build nested clusters by merging or splitting them successively by minimizing a linkage (distance) criteria
- Observations closer to each other (distance) can be connected to form a cluster
- They can be agglomerative or divisive
- Agglomerative algorithms use a bottom up approach starting with each sample observation X as a cluster and aggregates (merges) them successively into clusters
- Divisive use a top down approach starting with the entire sample as a cluster and successively dividing it into clusters
- It is represented as a hierarchical tree (dendrogram)
- Root is cluster of all samples and leaves are clusters of individual samples
- Different linkage criteria’s are:
- Single – minimizes distance between closest observations of pairs of clusters
- Average – minimizes average distance between all observations of pairs of clusters
- Complete (Maximum) – minimizes the maximum distance between observations of pairs of clusters
- Ward – minimizes the sum of squared differences within all clusters. Minimizes variance. Similar to k-means
- Single linkage while being the worst hierarchical strategy, works well for large datasets and can be computed efficiently
DBSCAN
- Density based clustering algorithms define clusters as areas of high density separated by areas of low density
- A cluster is a set of core samples close to each other (areas of high density) and a set of non-core samples close to a core sample (neighbors) but are not core samples themselves
- The closeness is calculated using a distance metric
- DBSCAN (Density-Based Spatial Clustering of Applications with Noise) is a popular density based algorithm
- It builds clusters using 2 parameters that define density
- min_samples – minimum no of samples required in a neighborhood for a core sample
- eps – maximum distance between two samples to be considered as each others’ neighbors
- Higher min_samples or lower eps indicates higher density to form a cluster
- Density based clusters can be of any shape (unlike k-means convex shape)
Gaussian Mixture Model
- Distribution based clustering algorithms build clusters with samples belonging to the same distribution
- Gaussian Mixture Model (GMM) is a popular method within them
- GMM is a probabilistic model that assumes that all data points are generated from a mixture of finite Gaussian distributions (normal distribution) with unknown parameters
- It implements the Expectation-Maximization algorithm, which finds the maximum-likelihood estimates of unknown parameters of the model which depends on latent variables (inferred from observed samples)
- They suffer from limitations like scalability
The results of the different algorithms show that KMeans gives the best results
- It is more widely used and has better interpretability
Plotting in higher dimensions (above 3D) is difficult so dimension reduction techniques like LDA and PCA can be used